StructuralGT Software
COMPASS develops an open-source package for the graph theoretic analysis of networked materials called StructuralGT. StructuralGT can operate on any images of networked materials, and has already been used to produce results on microscopy (confocal, atomic force, transmission electron, scanning electron) and macroscale videography in 2D, as well as 3D electron tomography.
Contributing to StructuralGT
We welcome all contributions to StructuralGT! Please feel free to open an issue or pull request on our GitHub.
Contributing to StructuralGT
We welcome all contributions to StructuralGT! Please feel free to open an issue or pull request on our GitHub.
Examples of StructuralGT
StructuralGT has been used for many pieces of research. Here are a few of our favorite examples.
Fast and Massive Production of Aramid Nanofibers via Molecule Intercalation

Cumulative spider plot of graph theoretical parameters of three different ANF aerogels prepared using traditional mechanical nanofibrillation with DMSO + KOH (left), nanofibrillation in DMSO + KOH + H2O (center), and molecule intercalation in DMSO + KOH + IPA (right) with significant structural differences being shown in the molecule-intercalated network.
Graph–Property Relationships for Complex Chiral Nanodendrimers
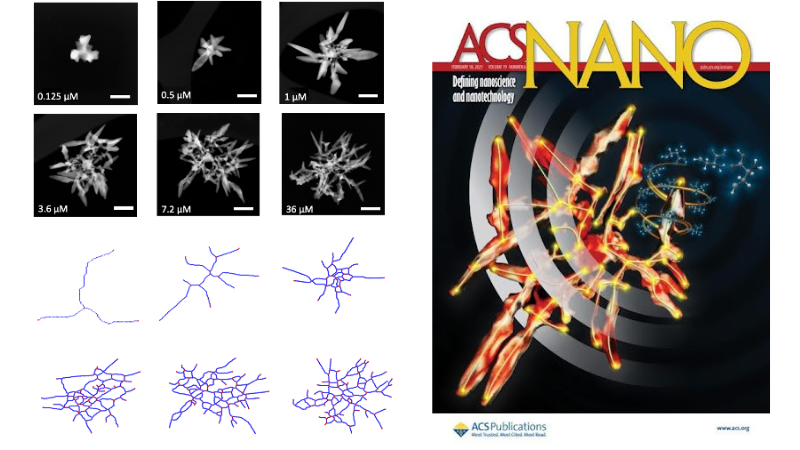
Left: Transmission electron microscopy of gold complex chiral gold nanodendrimers at different concentrations of cysteine, and their respective graphs. Right: Cover for the corresponding article.
Layer-by-layer assembled nanowire networks enable graph-theoretical design of multifunctional coatings
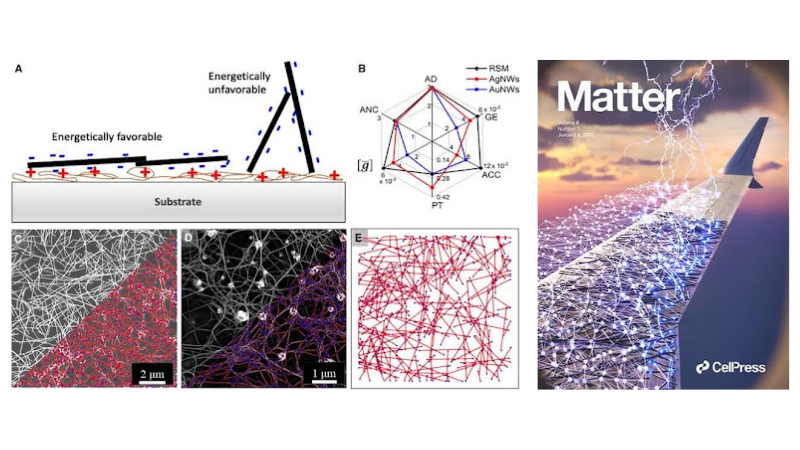
Left: Networks and graphs of silver NWs, gold NWs, and random stick models and their graph theoretical parameters. Right: Cover for the corresponding article.
Autocatalytic Nucleation and Self-Assembly of Inorganic Nanoparticles into Complex Biosimilar Networks
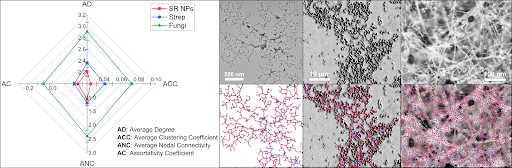
Comparative evaluation of biosimilarity of inorganic self-replicating nanoparticle (SR NP, left), bacterial (Streptococcus spp. on agar, center), and fungal (R. stolonifera on Capsicum annuum L, right) networks showing the NP networks are structurally identical to those produced by Streptococcus spp.
Beyond nothingness in the formation and functional relevance of voids in polymer films
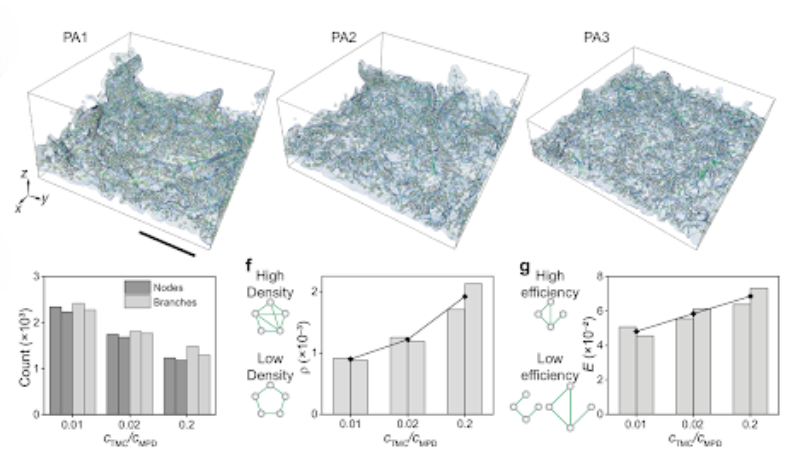
Top: Skeletonization schematics of tomography data of membrane voids. Bottom: Their graph theoretical parameters at different synthesis conditions.
